Conesa Lab releases bioinformatics software
A University of Florida Genetics Institute lab has created a bioinformatics web tool that detects microRNA binding elements in long non-coding RNA sequences. The software can be used to identify lncRNAs harboring multiple recognition elements of microRNAs.
The program is unique, as it is the only one that can be applied to all organisms, rather than strictly humans or mice, said Ana Conesa, a professor of microbiology & cell science and UFGI faculty member.
Conesa led the research team that created the software.
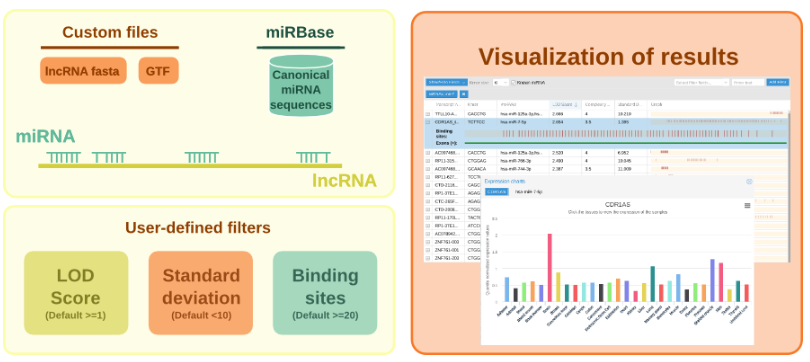
The program is called SpongeScan, and is available online.
Genomes have two types of genes– those that code for proteins, and those that do not. The latter are known as “non-coding RNA.” There are several subtypes of ncRNAs, and scientists are working to learn more about the roles of these genes in cell biology. Research indicates they play a role in regulating cellular processes.
MicroRNA, one subtype of non-coding RNA transcript, binds with RNA, in order to prevent it from creating a protein.
However, microRNAs also share sequence complementarity with lncRNAs, which enables the two to bind together.
When an lncRNA “absorbs” a microRNA, it prevents microRNA from binding to RNA, which leaves the RNA free to create a protein.
Conesa referred to this as the “sponge effect,” and said it signifies “a second layer of regulation of target protein levels.”
A few sponge lncRNAs have been identified in humans and mice, but it remains unclear how many of these molecules exist, and if they also occur in other organisms. The web tool created by Conesa’s team aims to help researchers in the discovery of novel sponges.
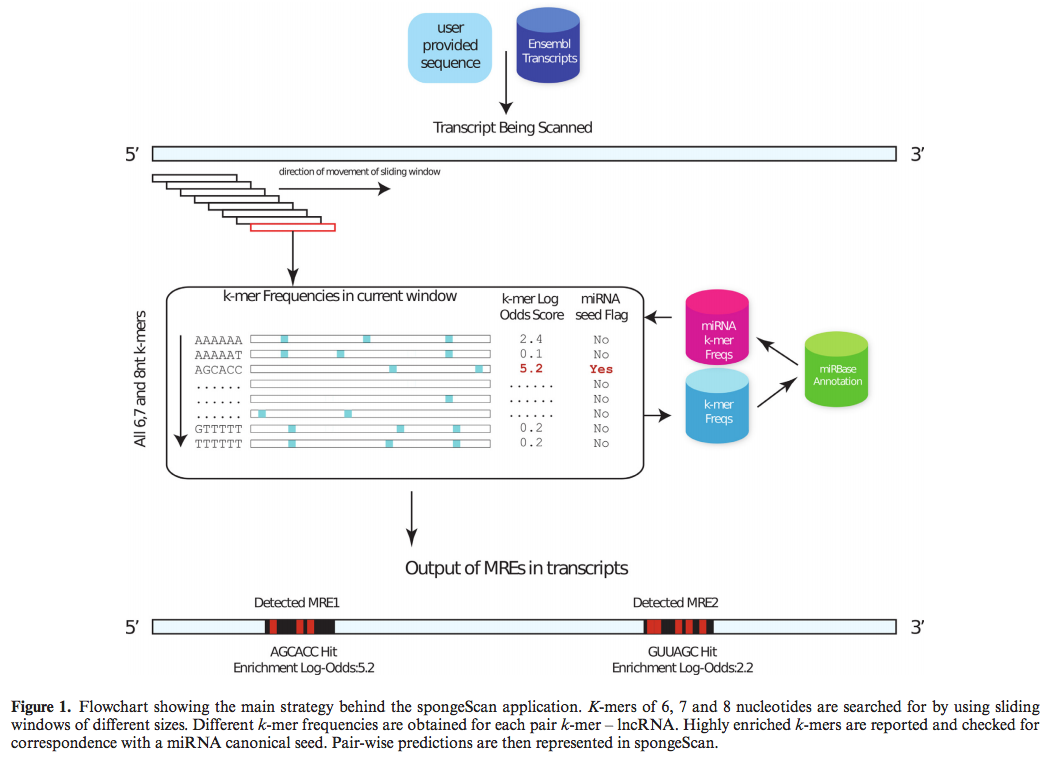
SpongeScan detects occurrences of distribution of specific MREs, and then ranks the pairings based on strength of compatibility.
The more binding sites that occur in a lncRNA sequence, the higher it will appear in the rankings as a potential sponge.
The program also returns information about number, position, conservation and sequence of biding sites; it is also able to analyze gene expression patterns of both lncRNAs and microRNAs to support their interaction, Conesa said.
Improving understanding of the sponge effect, and the binding patterns of molecules within cells, will help researchers better understand the role of novel genes for which the function is still unknown, Conesa said.